# 4 cifras significativas y sin notación científica:
options(digits = 4, scipen = 999)
# cargar librerías:
library(FactoClass)
library(plotly) # para gráficos interactivos
library(knitr) # para función kable (tablas estáticas)
library(DT) # para tablas interactivas
# cargar la tabla ejemplo admitidos:
data(admi)
n <- nrow(admi) # n: número de individuosEstadística descriptiva bivariada
Estadística Descriptiva Bivariada (Sección 3 del material de PyEF) https://cjtorresj.quarto.pub/pyef/03-estad_descr_bivar.html
Variables continuas
| mate | cien | soci | text | imag | |
|---|---|---|---|---|---|
| exam | 0.75 | 0.65 | 0.59 | 0.52 | 0.46 |
| age | 0.10 | -0.02 | -0.03 | -0.01 | 0.04 |
# Diagramas de dispersión y densidades _kernel_
plotpairs(admi[,2:6])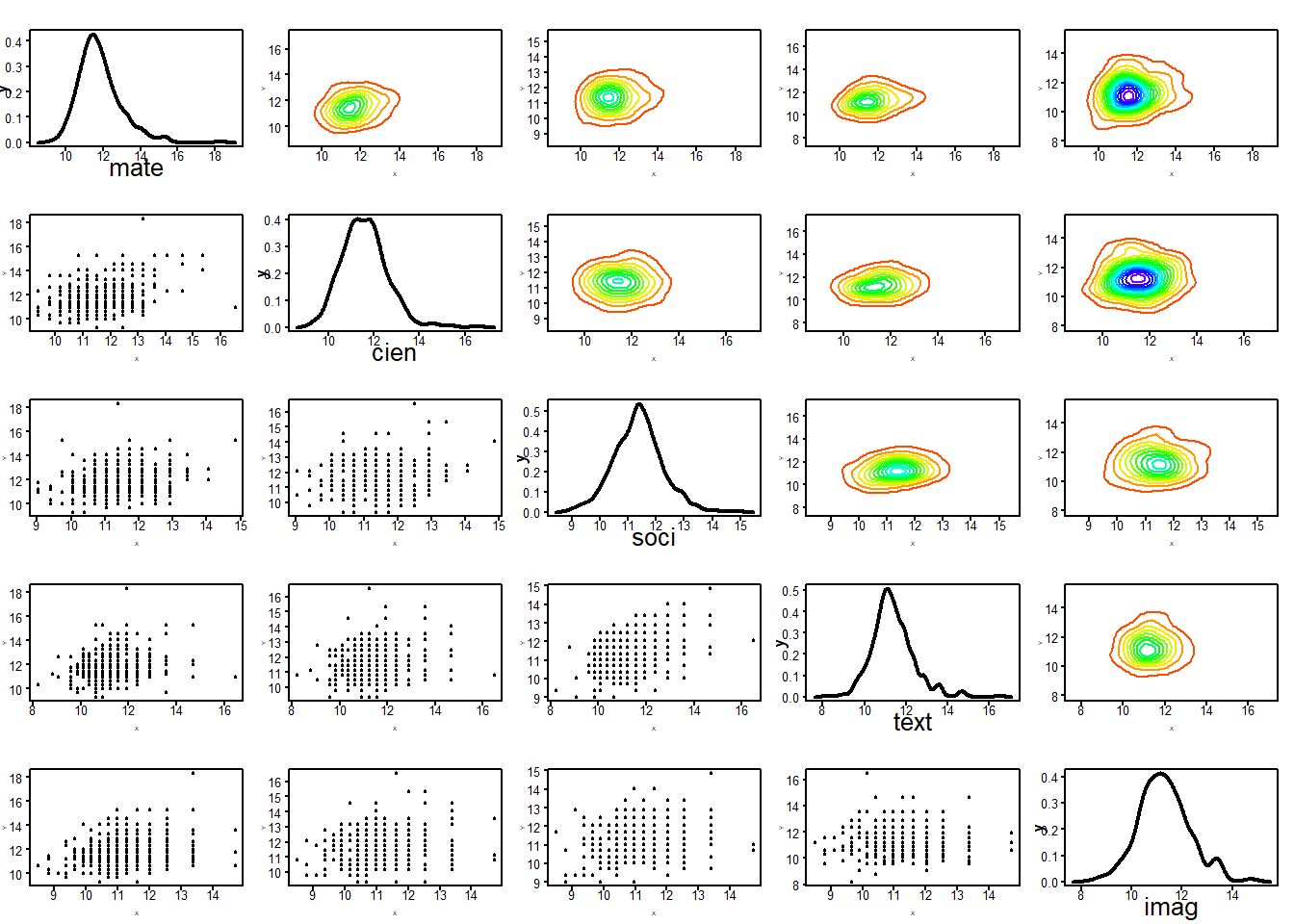
| mate | cien | soci | text | imag | |
|---|---|---|---|---|---|
| mate | 1.28 | 0.39 | 0.24 | 0.27 | 0.24 |
| cien | 0.39 | 1.00 | 0.14 | 0.20 | 0.12 |
| soci | 0.24 | 0.14 | 0.75 | 0.32 | 0.09 |
| text | 0.27 | 0.20 | 0.32 | 0.98 | 0.05 |
| imag | 0.24 | 0.12 | 0.09 | 0.05 | 1.01 |
Variables continua y cualitativa
# Diagramas de caja _boxplots_
par(bty="n")
sorted_carr <- with(admi, reorder(carr, mate, median, na.rm = T))
boxplot(admi$mate ~ sorted_carr, las = 1, col = 4, horizontal = T)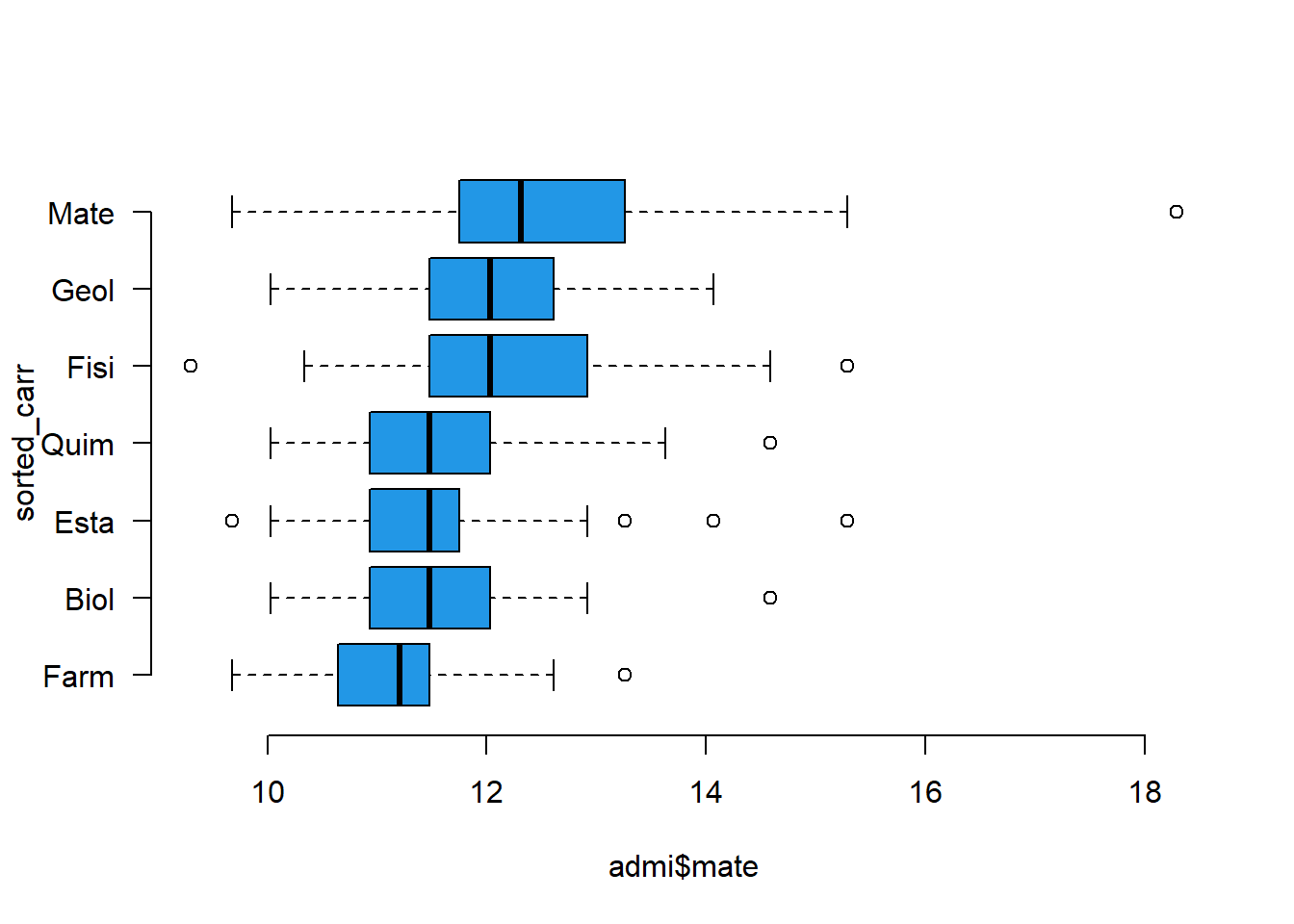
Razón de correlación
\eta^2_{XY} = \frac{\sigma^2_{entre}}{\sigma^2} donde \sigma^2_{entre} = \sum_{k=1}^{K} \frac{n_k}{n} \left( \bar{x}_k - \bar{x}\right)^2
# Gráficos de violín con puntos y media
sorted_carr <- with(admi, reorder(carr, mate, mean, na.rm=T))
fig <- admi %>% plot_ly(x = ~mate, split = sorted_carr, type = 'violin',
points = "all", pointpos = 0,
meanline = list(visible = T, color = "black"))
fig <- fig %>% layout(xaxis = list(title = "mate"),
yaxis = list(title = "sorted carr"))
figValores test
Sea X con E[X] = \mu y Var[X] = \sigma^2, entonces, para una muestra de tamaño n_k tomada con reemplazamiento (o para una población infinita), se tiene que: E[\bar{X}_k] = \mu y Var\left[\bar{X}_k\right] = \frac{\sigma^2}{n_k}.
Para una muestra sin reemplazamiento de una población finita n, se aplica el factor de corrección: \frac{n - n_k}{n - 1}, entonces, en este caso se tiene que: E[\bar{X}_k] = \mu y Var\left[\bar{X}_k\right] = \frac{n - n_k}{n - 1} \frac{\sigma^2}{n_k}.
Estandarizando el valor \bar{x}_k que tenemos, se obtiene que, \frac{\bar{x}_k - \mu}{\sigma_k} = \sqrt{\frac{(n - 1)n_k}{n - n_k}} \frac{\bar{x}_k - \mu}{\sigma}
Mediante la anterior fórmula podemos obtener valores indicativos, de qué tan lejos está una media \bar{x}_k con respecto a la media general o global, en términos de cantidad de desviaciones estándar \sigma_k, ya sea a la derecha o a la izquierda.
# Valores test para caracterización de carreras
# según los resultados del examen
cluster.carac(admi[,2:7], admi$carr, tipo.v = "co",
v.lim = 2, dn = 2, dm = 2, neg = TRUE)class: Biol
Test.Value Class.Mean Frequency Global.Mean
mate -2.26 11.5 63 11.8
------------------------------------------------------------
class: Esta
Test.Value Class.Mean Frequency Global.Mean
text -2.60 11.06 66 11.36
soci -2.84 11.08 66 11.36
cien -3.58 11.18 66 11.59
exam -3.74 680.21 66 718.35
------------------------------------------------------------
class: Farm
Test.Value Class.Mean Frequency Global.Mean
imag -2.94 10.98 73 11.3
exam -3.40 685.74 73 718.4
mate -4.47 11.25 73 11.8
------------------------------------------------------------
class: Fisi
Test.Value Class.Mean Frequency Global.Mean
mate 3.37 12.18 82 11.80
exam 3.32 748.01 82 718.35
cien 2.48 11.84 82 11.59
------------------------------------------------------------
class: Geol
Test.Value Class.Mean Frequency Global.Mean
exam 2.47 749.61 45 718.4
mate 2.04 12.12 45 11.8
------------------------------------------------------------
class: Mate
Test.Value Class.Mean Frequency Global.Mean
mate 5.82 12.65 53 11.8
exam 3.91 763.53 53 718.4
imag 3.13 11.70 53 11.3
------------------------------------------------------------
class: Quim
Test.Value Class.Mean Frequency Global.Mean
mate -2.1 11.52 63 11.8Variables cualitativas
# Tabla de contigencias edad - estrato (frec. abs. y rel.)
tc <- table(admi$edad, admi$estr)
tabtc <- cbind(tc, totF = rowSums(tc))
tabtc <- rbind(tabtc, totC=colSums(tabtc))
kable(cbind(tabtc, rep(" ", 5), round(tabtc/n*100, 1)),
digits = 1)| bajo | medio | alto | totF | bajo | medio | alto | totF | ||
|---|---|---|---|---|---|---|---|---|---|
| a16m | 44 | 47 | 27 | 118 | 9.9 | 10.6 | 6.1 | 26.5 | |
| a17 | 58 | 74 | 39 | 171 | 13 | 16.6 | 8.8 | 38.4 | |
| a18 | 22 | 26 | 8 | 56 | 4.9 | 5.8 | 1.8 | 12.6 | |
| a19M | 55 | 38 | 7 | 100 | 12.4 | 8.5 | 1.6 | 22.5 | |
| totC | 179 | 185 | 81 | 445 | 40.2 | 41.6 | 18.2 | 100 |
# Perfiles fila
pf <- prop.table(tc, 1)
kable(rbind(addmargins(pf, margin = 2)*100,
totC = tabtc[5,]/n*100), digits = 1)| bajo | medio | alto | Sum | |
|---|---|---|---|---|
| a16m | 37.3 | 39.8 | 22.9 | 100 |
| a17 | 33.9 | 43.3 | 22.8 | 100 |
| a18 | 39.3 | 46.4 | 14.3 | 100 |
| a19M | 55.0 | 38.0 | 7.0 | 100 |
| totC | 40.2 | 41.6 | 18.2 | 100 |
# Perfiles columna
pc <- prop.table(tc, 2)
kable(cbind(addmargins(pc, margin = 1)*100,
totF = tabtc[,4]/n*100), digits = 1)| bajo | medio | alto | totF | |
|---|---|---|---|---|
| a16m | 24.6 | 25.4 | 33.3 | 26.5 |
| a17 | 32.4 | 40.0 | 48.1 | 38.4 |
| a18 | 12.3 | 14.1 | 9.9 | 12.6 |
| a19M | 30.7 | 20.5 | 8.6 | 22.5 |
| Sum | 100.0 | 100.0 | 100.0 | 100.0 |
# Gráficos de los perfiles fila y perfiles columna
par(mfrow=c(2,1), mai=c(0.4,1,0.3,0.1))
plotct(t(tc), "row", col=2:5)
plotct(tc, "row", col=2:4)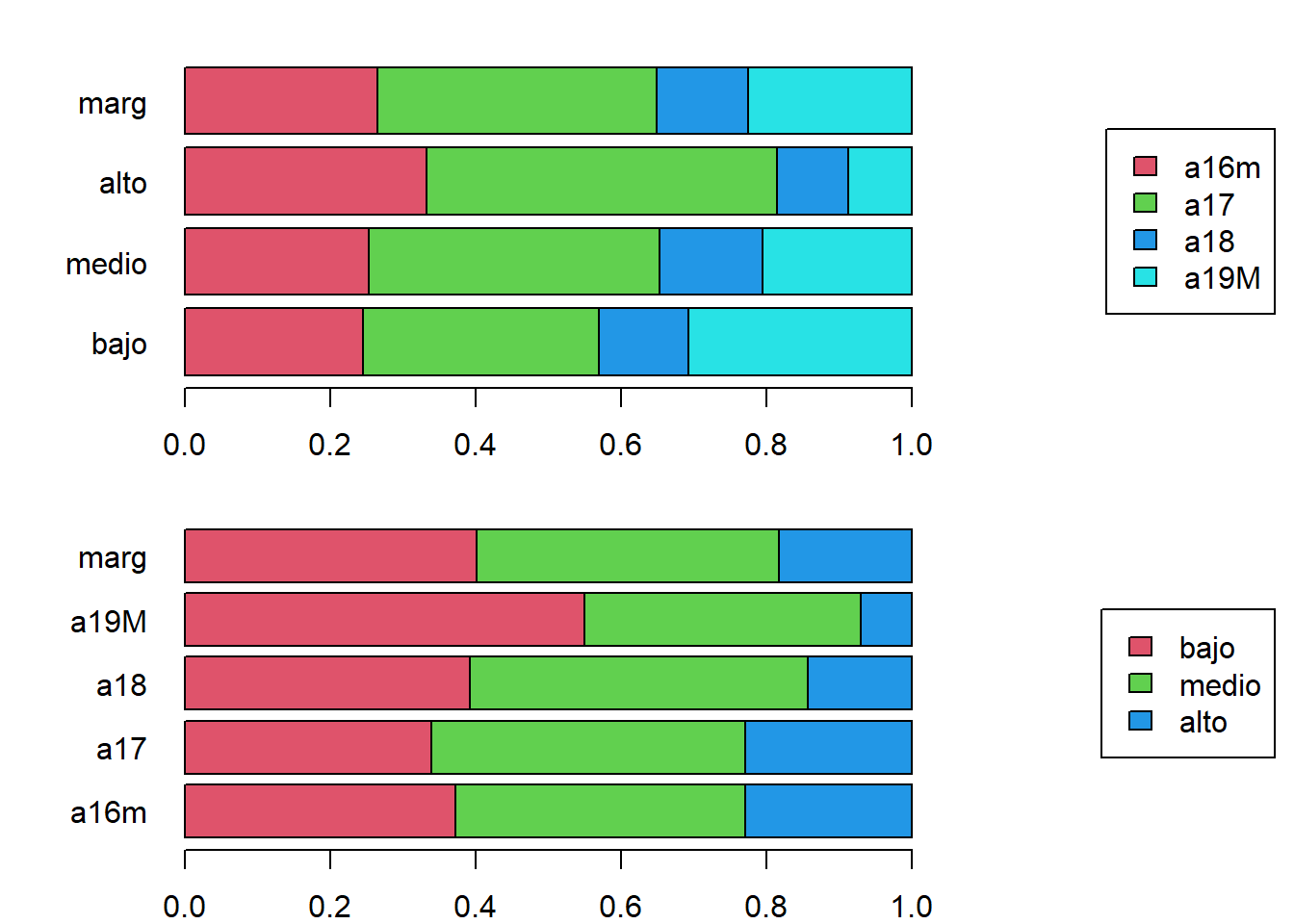
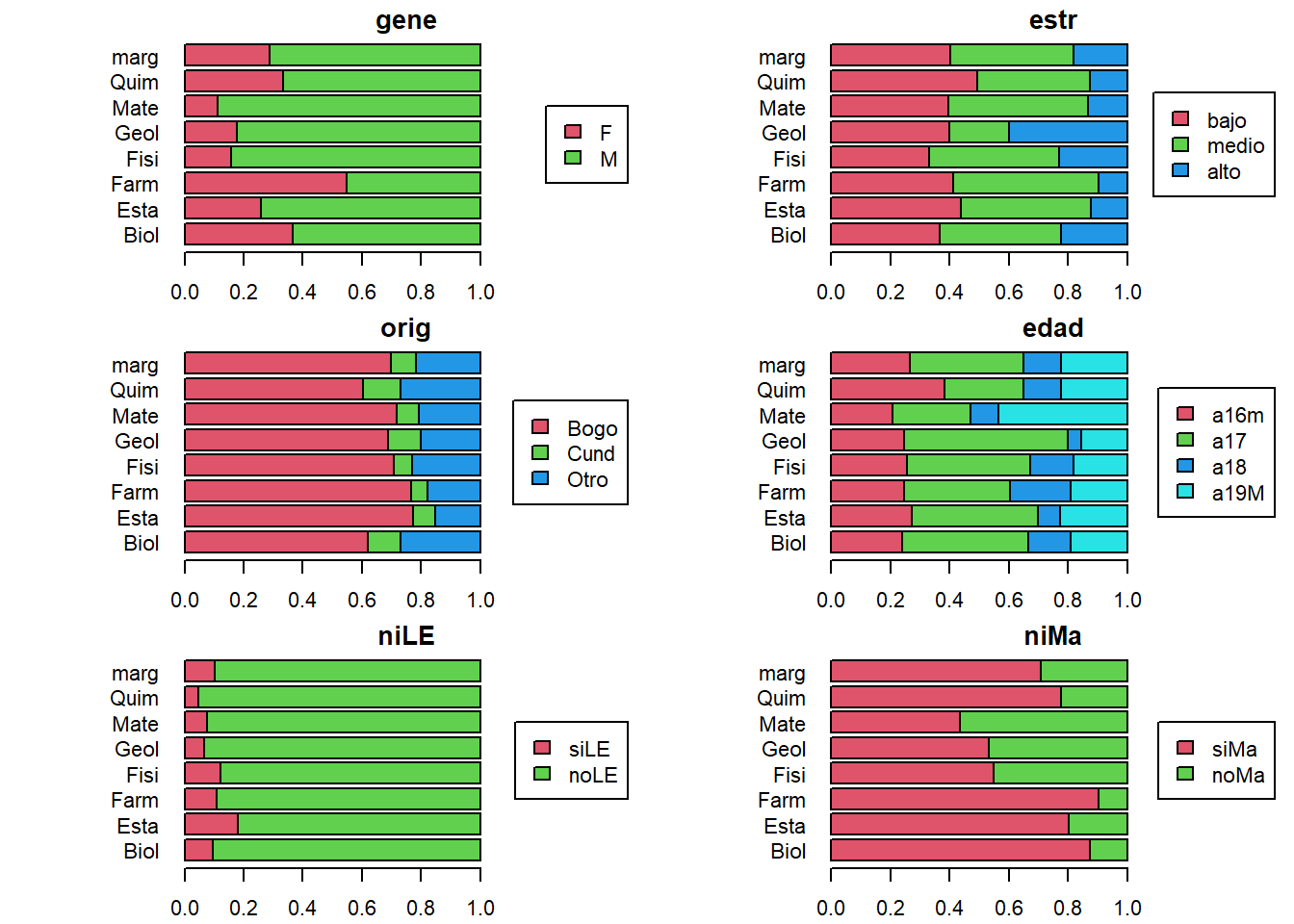
# Gráficos de perfiles de las carreras según cada cualitativa
par(mfrow = c(3,2), mai = c(0.3,1,0.2,0.1), las = 1, cex = 0.7)
for(i in 8:13){
tc <- unclass(table(admi$carr, admi[,i]))
plotct(tc, "row", col=1+(1:ncol(tc)))
title(main = names(admi)[i])
}
Medida de asociación
\chi^2 = \sum_{j=1}^J \sum_{k=1}^K \frac{\left(n_{jk}-\dfrac{n_{j\cdot} n_{\cdot k}}{n} \right)^2}{\dfrac{n_{j\cdot} n_{\cdot k}}{n} }
\phi^2 = \frac{\chi^2}{n}
# Valores indicativos de asociación
kable(chisq.carac(admi[,8:11], admi$carr, decr = FALSE), digits=3)| chi2 | dfr | pval | tval | phi2 | |
|---|---|---|---|---|---|
| gene | 44.108 | 6 | 0.000 | 5.264 | 0.099 |
| estr | 29.190 | 12 | 0.004 | 2.679 | 0.066 |
| orig | 9.676 | 12 | 0.644 | -0.370 | 0.022 |
| edad | 33.553 | 18 | 0.014 | 2.189 | 0.075 |
Valores test
# Tabla perfiles fila
kable(tabs$perR, digits = 1)| Biol | Esta | Farm | Fisi | Geol | Mate | Quim | |
|---|---|---|---|---|---|---|---|
| siMa | 17.5 | 16.8 | 21.0 | 14.3 | 7.6 | 7.3 | 15.6 |
| noMa | 6.2 | 10.0 | 5.4 | 28.5 | 16.2 | 23.1 | 10.8 |
| marg | 14.2 | 14.8 | 16.4 | 18.4 | 10.1 | 11.9 | 14.2 |
# Tabla perfiles columna
kable(tabs$perC, digits = 1)| Biol | Esta | Farm | Fisi | Geol | Mate | Quim | marg | |
|---|---|---|---|---|---|---|---|---|
| siMa | 87.3 | 80.3 | 90.4 | 54.9 | 53.3 | 43.4 | 77.8 | 70.8 |
| noMa | 12.7 | 19.7 | 9.6 | 45.1 | 46.7 | 56.6 | 22.2 | 29.2 |
# Tabla de contingencia (frec. rel.)
kable(tabs$ctm/n*100, digits = 1)| Biol | Esta | Farm | Fisi | Geol | Mate | Quim | marR | |
|---|---|---|---|---|---|---|---|---|
| siMa | 12.4 | 11.9 | 14.8 | 10.1 | 5.4 | 5.2 | 11.0 | 70.8 |
| noMa | 1.8 | 2.9 | 1.6 | 8.3 | 4.7 | 6.7 | 3.1 | 29.2 |
| marC | 14.2 | 14.8 | 16.4 | 18.4 | 10.1 | 11.9 | 14.2 | 100.0 |
# Tabla de contingencia (frec. abs.)
kable(tabs$ctm, digits = 0)| Biol | Esta | Farm | Fisi | Geol | Mate | Quim | marR | |
|---|---|---|---|---|---|---|---|---|
| siMa | 55 | 53 | 66 | 45 | 24 | 23 | 49 | 315 |
| noMa | 8 | 13 | 7 | 37 | 21 | 30 | 14 | 130 |
| marC | 63 | 66 | 73 | 82 | 45 | 53 | 63 | 445 |
# Valor test para Biología (k=1) y "siMa" (j=1)
# De n = 445, n_j = 315 tienen la característica "siMa"
# Al tomar una muestra sin reemplazamiento de n_k = 63
# ¿cuál es la probabilidad de que hayan salido n_kj = 55
# o algo "peor" (más alejado de la independencia)?
# valor p = P[ X >= 55 ] donde X ~ Hg(N = 445, M = 315, n = 63)
# P[ X >= 55 ] = 1 - P[ X <= 54 ]
(vp <- 1 - phyper(54, 315, 445-315, 63)) [1] 0.0008624# P[ X >= 55 ] = P[ X > 54 ]
(vp <- phyper(54, 315, 445-315, 63, lower.tail = FALSE)) [1] 0.0008624# Cuantil de la normal asociado a ese valor p (dos colas)
(t <- qnorm(vp/2, lower.tail = FALSE))[1] 3.332# Representación gráfica del cuantil (valor test) obtenido
plot(c(-5,5), c(-0.03, 0.4), type = "n", bty = "n",
axes = F, xlab = "", ylab = "")
abline(v = seq(-5,5), h = seq(0, 0.4, 0.1), col = "gray")
abline(v = c(-t,t), lty = 2, col = "red") # cuantiles
axis(1, pos=0)
axis(2, pos=0, las=1)
curve(dnorm(x), add = T, las = 1, col = "blue")
text(t, 0, signif(vp/2,2), adj = c(0,1), cex = 0.7, col = "blue")
text(t, -0.03, round(t,3), col = "red", cex = 0.9)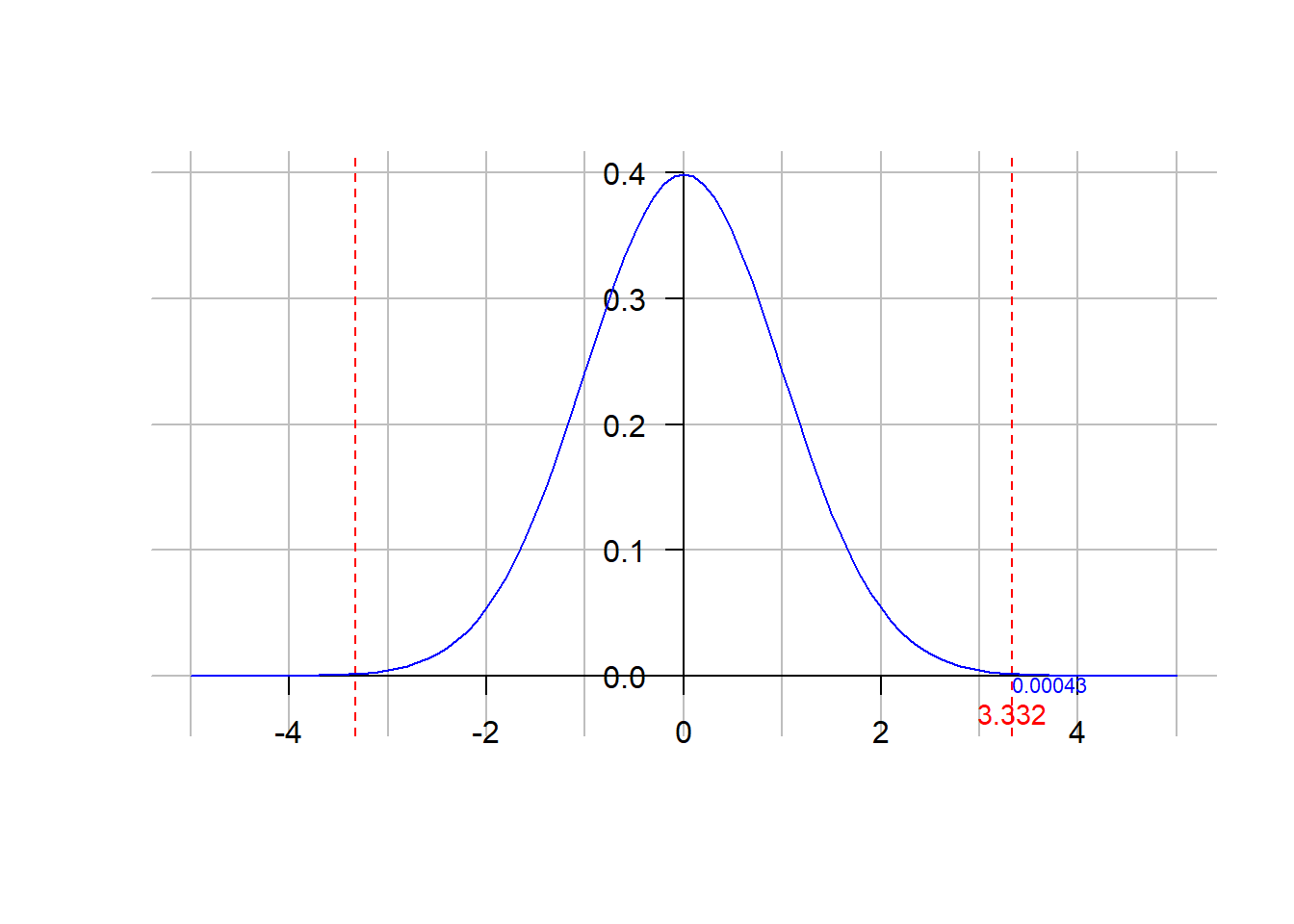
# Valores test para caracterización de carreras
# según las variables cualitativas
cluster.carac(admi[,8:13], admi[,1])class: Biol
Test.Value p.Value Class.Cat Cat.Class Global Weight
niMa.siMa 3.332 0.001 17.5 87.3 70.8 315
niMa.noMa -3.332 0.001 6.2 12.7 29.2 130
------------------------------------------------------------
class: Esta
Test.Value p.Value Class.Cat Cat.Class Global Weight
niLE.siLE 2.235 0.025 26.1 18.2 10.3 46
niMa.siMa 2.034 0.042 16.8 80.3 70.8 315
niMa.noMa -2.034 0.042 10.0 19.7 29.2 130
niLE.noLE -2.235 0.025 13.5 81.8 89.7 399
------------------------------------------------------------
class: Farm
Test.Value p.Value Class.Cat Cat.Class Global Weight
gene.F 5.152 0.000 31.2 54.8 28.8 128
niMa.siMa 4.355 0.000 21.0 90.4 70.8 315
edad.a18 2.252 0.024 26.8 20.5 12.6 56
estr.alto -2.281 0.023 8.6 9.6 18.2 81
niMa.noMa -4.355 0.000 5.4 9.6 29.2 130
gene.M -5.152 0.000 10.4 45.2 71.2 317
------------------------------------------------------------
class: Fisi
Test.Value p.Value Class.Cat Cat.Class Global Weight
niMa.noMa 3.475 0.001 28.5 45.1 29.2 130
gene.M 3.045 0.002 21.8 84.1 71.2 317
gene.F -3.045 0.002 10.2 15.9 28.8 128
niMa.siMa -3.475 0.001 14.3 54.9 70.8 315
------------------------------------------------------------
class: Geol
Test.Value p.Value Class.Cat Cat.Class Global Weight
estr.alto 3.677 0.000 22.2 40.0 18.2 81
niMa.noMa 2.706 0.007 16.2 46.7 29.2 130
edad.a17 2.554 0.011 14.6 55.6 38.4 171
niMa.siMa -2.706 0.007 7.6 53.3 70.8 315
estr.medio -3.242 0.001 4.9 20.0 41.6 185
------------------------------------------------------------
class: Mate
Test.Value p.Value Class.Cat Cat.Class Global Weight
niMa.noMa 4.467 0.000 23.1 56.6 29.2 130
edad.a19M 3.683 0.000 23.0 43.4 22.5 100
gene.M 3.218 0.001 14.8 88.7 71.2 317
edad.a17 -2.089 0.037 8.2 26.4 38.4 171
gene.F -3.218 0.001 4.7 11.3 28.8 128
niMa.siMa -4.467 0.000 7.3 43.4 70.8 315
------------------------------------------------------------
class: Quim
Test.Value p.Value Class.Cat Cat.Class Global Weight
edad.a16m 2.320 0.020 20.3 38.1 26.5 118
edad.a17 -2.189 0.029 9.9 27.0 38.4 171Taller (razas de perros)
Caracterización de la función de razas de perros
data(DogBreeds) # cargar datos
# Tabla dinámica con función datatable {DT}:
datatable(DogBreeds, style = "bootstrap4",
options = list(pageLength = 5))summary(DogBreeds) # Resumen SIZE WEIG SPEE INTE AFFE AGGR FUNC
lar:15 hea: 5 hig: 9 hig: 6 hig:14 hig:13 com:10
med: 5 lig: 8 low:10 low: 8 low:13 low:14 hun: 9
sma: 7 med:14 med: 8 med:13 uti: 8 # chisq.carac (valores indicativos de asociación)
kable(chisq.carac(DogBreeds[,1:6], DogBreeds[,7],
decr = FALSE),
digits=3)| chi2 | dfr | pval | tval | phi2 | |
|---|---|---|---|---|---|
| SIZE | 16.354 | 4 | 0.003 | 2.797 | 0.606 |
| WEIG | 24.407 | 4 | 0.000 | 3.822 | 0.904 |
| SPEE | 8.483 | 4 | 0.075 | 1.437 | 0.314 |
| INTE | 4.140 | 4 | 0.387 | 0.286 | 0.153 |
| AFFE | 14.761 | 2 | 0.001 | 3.228 | 0.547 |
| AGGR | 7.073 | 2 | 0.029 | 1.894 | 0.262 |
# cluster.carac (valores test)
cluster.carac(DogBreeds[,1:6], DogBreeds[,7], dn = 2, dm = 2)class: com
Test.Value p.Value Class.Cat Cat.Class Global Weight
AFFE.hig 3.85 0 71.4 100 51.9 14
WEIG.lig 3.31 0 87.5 70 29.6 8
SIZE.sma 2.87 0 85.7 60 25.9 7
SIZE.lar -3.54 0 6.7 10 55.6 15
AFFE.low -3.85 0 0.0 0 48.1 13
------------------------------------------------------------
class: hun
Test.Value p.Value Class.Cat Cat.Class Global Weight
WEIG.med 2.62 0.01 57.1 88.9 51.9 14
AFFE.low 2.08 0.04 53.8 77.8 48.1 13
AFFE.hig -2.08 0.04 14.3 22.2 51.9 14
------------------------------------------------------------
class: uti
Test.Value p.Value Class.Cat Cat.Class Global Weight
WEIG.hea 3.39 0.00 100.0 62.5 18.5 5
SIZE.lar 2.98 0.00 53.3 100.0 55.6 15
AGGR.hig 2.53 0.01 53.8 87.5 48.1 13
WEIG.lig -2.12 0.03 0.0 0.0 29.6 8
SPEE.med -2.12 0.03 0.0 0.0 29.6 8
AGGR.low -2.53 0.01 7.1 12.5 51.9 14